COVID-19 Research at NERSC
In the early stage of the COVID-19 pandemic NERSC quickly provided urgent access to its High Performance Computing and Data resources and staff expertise. Projects got access to NERSC to conduct COVID-related research through a number of different means, including NERSC's standard mission science ("ERCAP") allocation process and through strategic collaborations between NERSC/Berkeley Lab and research teams.
In addition to ERCAP and strategic engagements, NERSC also participated in two programs that provided support for COVID research, but are no longer accepting new COVID projects: the National COVID-19 HPC Consortium and awards from the C3.ai Digital Transformation Institute. Some projects were also funded by the DOE Exascale Computing Project (ECP).
NERSC continues to support groundbreaking research in this area, resulting in a number of scientific publications as shown in this sample of published research in 2022:
- "Structural Insights into Binding of Remdesivir Triphosphate within the Replication-Transcription Complex of SARS-CoV-2"; Biochemistry, 61:1966-1973; 2022 Aug 31, 10.1021/acs.biochem.2c00341
- "Translocation pause of remdesivir-containing primer/template RNA duplex within SARS-CoV-2's RNA polymerase complexes"; Frontiers in Molecular Biosciences, 9:; 2022 Oct 25, 10.3389/fmolb.2022.999291
- "Computational Design of Miniproteins as SARS-CoV-2 Therapeutic Inhibitors"; International Journal of Molecular Sciences, 23:838; 2022 Jan 13, 10.3390/ijms23020838
- "Effect of Delta and Omicron Mutations on the RBD-SD1 Domain of the Spike Protein in SARS-CoV-2 and the Omicron Mutations on RBD-ACE2 Interface Complex"; International Journal of Molecular Sciences, 23:10091; 2022 Sep 6, 10.3390/ijms231710091
- "Molecular insights into the differential dynamics of SARS-CoV-2 variants of concern"; Journal of Molecular Graphics and Modelling, 114:108194; 2022 Apr 14, 10.1016/j.jmgm.2022.108194
- "MoS2 nanosheets effectively bind to the receptor binding domain of the SARS-CoV-2 spike protein and destabilize the spike-human ACE2 receptor interactions"; Soft Matter, 18:8961-8973; 2022 Oct 20, 10.1039/d2sm01181f
- "How Does Temperature Affect the Dynamics of SARS-CoV-2 M Proteins? Insights from Molecular Dynamics Simulations"; The Journal of Membrane Biology, 255:341-356; 2022 May 13, 10.1007/s00232-022-00244-y
- "Identification and mechanistic basis of non-ACE2 blocking neutralizing antibodies from COVID-19 patients with deep RNA sequencing and molecular dynamics simulations"; Frontiers in Molecular Biosciences, 9:; 2022 Dec 16, 10.3389/fmolb.2022.1080964

Viral Fate & Transport for COVID-19
Potentially infectious sites are created when COVID-19 carrying aerosol particles are released as a person talks/sings/coughs/sneezes/breathes and the virus is deposited on the ground and other surfaces. This team is studying how this process occurs in a model classroom setting.
Read More »

Computer-Aided Design of Peptide Ligands to SARS-CoV-2 Targets
Researchers from the Beckman Institute at the City of Hope are designing peptides and small molecules that will bind to the coronavirus surface proteins and inhibit their binding to human proteins. Besides potential therapy, these inhibitory peptides and molecules are useful for understanding the mechanisms of virus entry and interaction with the human host and the immune system. Read More »

Manipulation of Membrane Curvature by Designer Peptides for Battling COVID-19 Infection
Scientists from Boston University and UCLA are working closely to understand the factors (sequence and membrane composition) that dictate the activity of SARS-CoV-2 "inverse translocation peptides" and their effect on the structure of the human cell membranes . Read More »

NERSC & LCLS Team Up on SARS-CoV-2 Research
NERSC his working with scientists and staff at LCLS to enable real-time data analysis for two LCLS experiments looking at the structure of the SARS-CoV-2 virus. Read More »
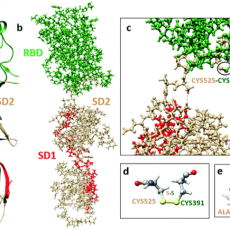
Structure Refinement of the Spike Protein of SARS-COV-2
University of Missouri, Kansas City scientists are refining the structure of regions on the SARS-COV-2 to much higher accuracy using large scale ab initio simulations and investigating their atomic scale interactions and intra molecular binding mechanisms. Read More »
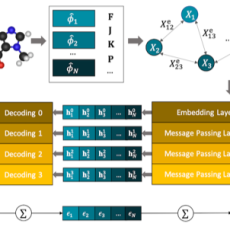
Machine Learning to model the full space of chemical biology and drug discovery with quantum-mechanical accuracy
This project is using their new machine learning (ML) model to screen drug molecules for an affinity with protein targets associated with COVID-19 infection. The goal is to span the full space of chemical biology and drug discovery with quantum-mechanical accuracy at much reduced computational cost.
Read More »
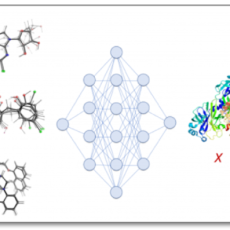
Drug-Repurposing for Covid-19 with 3D-Aware Machine Learning
Scientists from MIT and Harvard are exploring whether the repurposing of drugs for COVID-19 treatment can be accelerated with a combination of physical simulation and machine learning (ML). Read More »
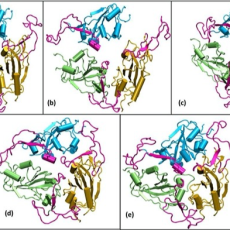
Dependence of Structure and Dynamics of Novel SARS-CoV-2 on Temperature and Humidity in the Atmosphere
Researchers from National Institute of Technology Warangal are studying whether the virus undergoes any biophysical changes in response to changes in atmospheric conditions, mainly temperature and humidity. Read More »