NERSC Supercomputers Help Berkeley Lab Scientists Map Key DNA Protein Complex
Cryo-electron microscopy aids gene research and drug development
September 13, 2017
Contact: (510) 486-4575
Chalking up another success for a new imaging technology that has energized the field of structural biology, researchers at the Department of Energy’s Lawrence Berkeley National Laboratory (Berkeley Lab) obtained the highest resolution map yet of a large assembly of human proteins that is critical to DNA function.
The scientists reported their achievement today in an advance online publication of the journal Nature. They used cryo-electron microscopy (cryo-EM) to resolve the 3-D structure of a protein complex called transcription factor IIH (TFIIH) at 4.4 angstroms, or near-atomic resolution. This protein complex is used to unzip the DNA double helix so that genes can be accessed and read during transcription or repair.
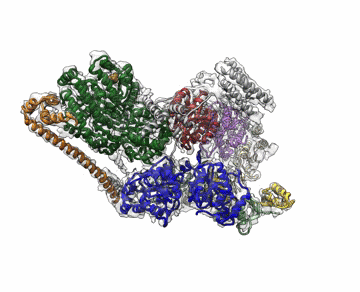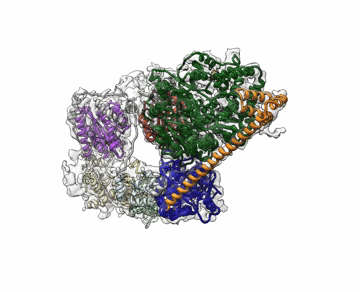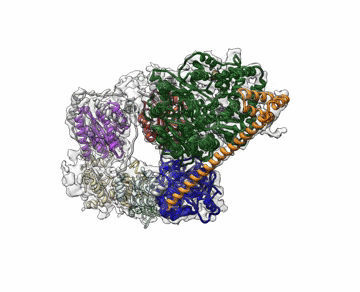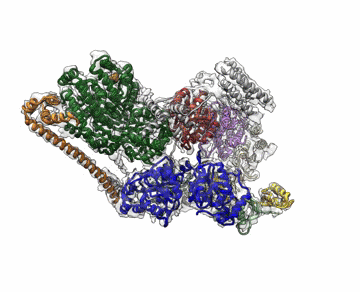
The cryo-EM structure of Transcription Factor II Human (TFIIH). The atomic coordinate model, colored according to the different TFIIH subunits, is shown inside the semi-transparent cryo-EM map. Credit: Basil Greber/Berkeley Lab and UC Berkeley
“When TFIIH goes wrong, DNA repair can’t occur, and that malfunction is associated with severe cancer propensity, premature aging, and a variety of other defects,” said study principal investigator Eva Nogales, faculty scientist at Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging Division. “Using this structure, we can now begin to place mutations in context to better understand why they give rise to misbehavior in cells.”
TFIIH’s critical role in DNA function has made it a prime target for research, but it is considered a difficult protein complex to study, especially in humans.
Mapping complex proteins
“As organisms get more complex, these proteins do, too, taking on extra bits and pieces needed for regulatory functions at many different levels,” said Nogales, who is also a UC Berkeley professor of molecular and cell biology and a Howard Hughes Medical Institute investigator. “The fact that we resolved this protein structure from human cells makes this even more relevant to disease research. There’s no need to extrapolate the protein’s function based upon how it works in other organisms.”
Biomolecules such as proteins are typically imaged using X-ray crystallography, but that method requires a large amount of stable sample for the crystallization process to work. The challenge with TFIIH is that it is hard to produce and purify in large quantities, and once obtained, it may not form crystals suitable for X-ray diffraction.
Enter cryo-EM, which can work even when sample amounts are very small. Electrons are sent through purified samples that have been flash-frozen at ultracold temperatures to prevent crystalline ice from forming.
Cryo-EM has been around for decades, but major advances over the past five years have led to a quantum leap in the quality of high-resolution images achievable with this technique.
“When your goal is to get resolutions down to a few angstroms, the problem is that anymotion gets magnified,” said study lead author Basil Greber, a UC Berkeley postdoctoral fellow at the California Institute for Quantitative Biosciences (QB3). “At high magnifications, the slight movement of the specimen as electrons move through leads to a blurred image.”
Making movies
The researchers credit the explosive growth in cryo-EM to advanced detector technology that Berkeley Lab engineer Peter Denes helped develop. Instead of a single picture taken for each sample, the direct detector camera shoots multiple frames in a process akin to recording a movie. The frames are then put together to create a high-resolution image. This approach resolves the blur from sample movement. The improved images contain higher quality data, and they allow researchers to study the sample in multiple states, as they exist in the cell.
Since shooting a movie generates far more data than a single frame, and thousands of movies are being collected during a microscopy session, the researchers needed the processing punch of supercomputers at the National Energy Research Scientific Computing Center (NERSC) at Berkeley Lab. The output from these computations was a 3-D map that required further interpretation.
“When we began the data processing, we had 1.5 million images of individual molecules to sort through,” said Greber. “We needed to select particles that are representative of an intact complex. After 300,000 CPU hours at NERSC, we ended up with 120,000 images of individual particles that were used to compute the 3-D map of the protein.”
To obtain an atomic model of the protein complex based on this 3-D map, the researchers used PHENIX (Python-based Hierarchical ENvironment for Integrated Xtallography), a software program whose development is led by Paul Adams, director of Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging Division and a co-author of this study.
Not only does this structure improve basic understanding of DNA repair, the information could be used to help visualize how specific molecules are binding to target proteins in drug development.
“In studying the physics and chemistry of these biological molecules, we’re often able to determine what they do, but how they do it is unclear,” said Nogales. “This work is a prime example of what structural biologists do. We establish the framework for understanding how the molecules function. And with that information, researchers can develop finely targeted therapies with more predictive power.”
Other co-authors on this study are Pavel Afonine and Thi Hoang Duong Nguyen, both of whom have joint appointments at Berkeley Lab and UC Berkeley; and Jie Fang, a researcher at the Howard Hughes Medical Institute.
NERSC is a DOE Office of Science User Facility located at Berkeley Lab. In addition to NERSC, the researchers used the Lawrencium computing cluster at Berkeley Lab. This work was funded by the National Institute of General Medical Sciences and the Swiss National Science Foundation.
RELATED STORIES
About NERSC and Berkeley Lab
The National Energy Research Scientific Computing Center (NERSC) is a U.S. Department of Energy Office of Science User Facility that serves as the primary high performance computing center for scientific research sponsored by the Office of Science. Located at Lawrence Berkeley National Laboratory, NERSC serves almost 10,000 scientists at national laboratories and universities researching a wide range of problems in climate, fusion energy, materials science, physics, chemistry, computational biology, and other disciplines. Berkeley Lab is a DOE national laboratory located in Berkeley, California. It conducts unclassified scientific research and is managed by the University of California for the U.S. Department of Energy. »Learn more about computing sciences at Berkeley Lab.







