Theoretical Study on Catalysis by Protein Enzymes and Ribozyme
2000 NERSC Annual Report
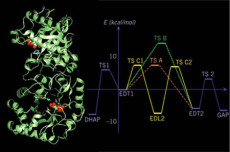
The energetics were determined for three mechanisms proposed for TIM catalyzed reactions. Results from reaction path calculations suggest that the two mechanisms that involve an enediol intermediate are likely to occur, while the direct intra-substrate proton transfer mechanism (in green) is energetically unfavorable due to the presence of His95 in the active site.
Principal Investigator: Martin Karplus, Harvard University
Research Objectives
The goal of this project is to develop a greater understanding of the mechanisms involved in enzyme catalysis and related protein functions. We are studying two types of enzymes: proteins and a nucleic acid (hammerhead ribozyme).
Computational Approach
For active-site models in the gas phase, ab initio or density functional (DFT) calculations are used. A few calculations with continuum dieletric models are carried out to investigate the effect of solvation; the results are compared with those in the enzyme. Those calculations are carried out using mainly Gaussian98 and NWChem. To determine the catalytic mechanism in the presence of the enzyme environment, a combined quantum/molecular mechanics (QM/MM) approach is used, performed with the CHARMM program. To study the effect of tunneling on proton or hydride transfer, variational transition state theory is used.
Accomplishments
Hammerhead ribozymes: Ab initio and DFT calculations have been carried out to study the reaction path in the phosphate ester hydrolysis of an RNA model which represents a minimum active site of the hammerhead ribozyme. Our results help explain the loss of catalytic activity observed experimentally when replacing bridging or non-bridging oxygen atoms from the phosphate group.
Yeast chorismate mutase (YCM): Preliminary calculations have been carried out for the rearrangement of chorismate to prephenate in the gas phase and in solution. Optimized structures for several comformations have been obtained from ab initio and DFT calculations. Our calculations identified a pathway for the elimination reaction from chorismate to 4-hydroxybenzoate and explained the experimental observation that the relative rates of the rearrangement and elimination reactions depend on solvents.
Triosephosphate isomerase (TIM): Three catalytic mechanisms proposed in the literature were studied with the combined DFT/MM approach. The two pathways that involve an enediol species were found to be give similar values for the barriers, in satisfactory agreement with experiment. The mechanism that involves intramolecular proton transfer in the enediolate was found to be energetically unfavorable due to the presence of His95. We also applied variational transition state theory (VTST) to investigate the effect of tunneling on two proton transfer steps in TIM. It was found that tunneling has a significant but not very large effect on the rate constants at room temperature, and appears to be consistently more significant in enzyme than for the corresponding reaction in solution.
Significance
Details of the chemical mechanisms employed by enzymes to serve as catalysts of biochemical reactions remain elusive, largely because the chemical events of bond formation and cleavage are exceedingly short and are currently inaccessible to direct experimental measurement. Theoretical studies, therefore, provide valuable insights into enzyme catalysis.
Publications
P. D. Lyne and M. Karplus, "Determination of the pKa of the 2'-hydroxyl group at the active site of hammerhead ribozyme from ab initio calculations with solvation corrections," J. Am. Chem. Soc. 122, 166 (2000).
Q. Cui and M. Karplus, "Molecular properties from combined QM/MM methods. I. Analytical second derivative and vibrational calculations," J. Chem. Phys. 112, 1133 (2000).
Q. Cui and M. Karplus, "Molecular properties from combined QM/MM methods. II. Chemical shifts in large molecules," J. Phys. Chem. B 104, 3721 (2000).
About NERSC and Berkeley Lab
The National Energy Research Scientific Computing Center (NERSC) is a U.S. Department of Energy Office of Science User Facility that serves as the primary high performance computing center for scientific research sponsored by the Office of Science. Located at Lawrence Berkeley National Laboratory, NERSC serves almost 10,000 scientists at national laboratories and universities researching a wide range of problems in climate, fusion energy, materials science, physics, chemistry, computational biology, and other disciplines. Berkeley Lab is a DOE national laboratory located in Berkeley, California. It conducts unclassified scientific research and is managed by the University of California for the U.S. Department of Energy. »Learn more about computing sciences at Berkeley Lab.







