Nano-Behavior
SCIENTISTS USE SIMULATIONS TO STUDY DNA INTERACTIONS WITH PROTEINS, POLYMER
March 1, 2008
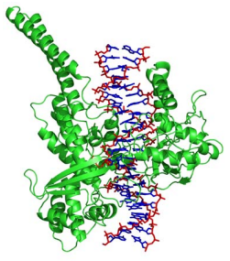
The image shows the relationship between Topoisomerase and DNA. Topoisomerase (in green) is an enzyme that unknots the DNA, allowing proteins to transcribe and form a new set of genetic materials during cell division.
A team from the University of California at Irvine is exploring the complex interactions of human DNA with proteins and nanoparticles in a research that tackles key problems in designing and delivering anti-cancer drugs and gene therapy.
The research team, headed by Ioan Andricioaei, is using the computer power and services at NERSC to better understand how certain proteins affect DNA’s behavior during cell replication. The research also looks at the dynamics between DNA and polyamidoamine (PAMAM) dendrimers, nano-particles that could be used to transport drugs and genetic materials into the body for treatment.
The research builds on previous molecular dynamic simulations by the team. The 2008 allocation of 7.4 million MPP hours represents the first major undertaking at NERSC by the Irvine research team.
“Our simulations require thousands of processors to simulate the vast conformational changes we are attempting to study,” says Jeff Wereszczynski of the Andricioaei team. “With the power of Franklin (Cray XT4), what used to take us several months we can now do in weeks.”
In the first project, the scientists want to know how an enzyme called topoisomerase I, which loosens the tightly packed DNA during replication, would react to topotecan. Topotecan is a chemical compound known to disrupt the enzyme’s work, and is currently used for treating ovarian and lung cancer. But exactly how topoisomerase I and DNA interact with topotecan isn’t clear. Understanding their dynamics could lead to better-designed cancer treatments.
The research conducted by Wereszczynski looks specifically at the energy use and movement of DNA as supercoils are relaxed, a vital step to allow cell division, and at how topotecan affects this function. Previous simulation work by the team already showed a difference in the mechanisms at which twisted DNA strands relax without interference from topotecan, so that proteins can access genetic information and jumpstart the replication process (Sari and Andricioaei, Nucleic Acids Research, 2005).
In the second project, Andricioaei and Mills aim to figure out how DNA behaves when it’s attached to a nano-particle such as PAMAM dendrimer. Does the pairing alter the structure or properties of both objects, for example? Does it matter what part of the PAMAM dendrimer surface is used?
PAMAM dendrimers are tiny, and a variety of molecules can be easily attached to their surface. For this reason, PAMAM dendrimers are considered promising vehicles for delivering healthy DNA to fix faulty genes.
Based on their previous work, the scientists knew that both the DNA and the dendrimer would become deformed if the widest edge of a PAMAM dendrimer is parallel to the axis of a DNA. The DNA would wrap around the dendrimer while the dendrimer would flatten against the DNA. Understanding the energetics and dynamics behind this interaction is key in advancing the nanotechnology uinderlying gene therapies further.
In the third project, the researchers seek to understand the role of portal pro- tein in the life of a bacteriophage, which infects a bacterium by pushing its genetic material into the host’s cells. Since the early 1900s, scientists have discovered that some viruses can act as parasites and kill their bacterial hosts. Since antibi- otics are easy to manufacture and store, the idea of using bacteriophages to treat illnesses didn’t catch on, except in the former Soviet Union and Eastern Europe.
Understanding how bacteriophages work can lead to better medicines, including countering the effects of bioweapons.
What happens after a bacteriophage injects its DNA into a bacterial cell is the focus of the simulation performed by Jeremiah Nummela in the Andricioaei group. A bacteriophage consists of a protein shell enclosing its genetic material. When the viral DNA hijacks the protein synthesis mechanisms of the bacterium and begins to replicate itself, the process also involves the forming of capsids. Capsids are protein shells that would house these new DNA strands and become new bacteriophages.
The DNA strands enter the new capsids through the portal proteins. Importing the genetic materials through the portal proteins and packing them tightly requires a lot of force. Through simulations, the researchers intend to calculate the interaction between the DNA and portal protein. Andricioaei and Nummela will use a new method they have developed to carry out the calculations (Nummela and Andricioaei, Biophysical Journal, 2007). Learn more about the research at Andricioaei’s web site.
About NERSC and Berkeley Lab
The National Energy Research Scientific Computing Center (NERSC) is a U.S. Department of Energy Office of Science User Facility that serves as the primary high performance computing center for scientific research sponsored by the Office of Science. Located at Lawrence Berkeley National Laboratory, NERSC serves almost 10,000 scientists at national laboratories and universities researching a wide range of problems in climate, fusion energy, materials science, physics, chemistry, computational biology, and other disciplines. Berkeley Lab is a DOE national laboratory located in Berkeley, California. It conducts unclassified scientific research and is managed by the University of California for the U.S. Department of Energy. »Learn more about computing sciences at Berkeley Lab.







