Protein Dynamics and Biocatalysis
1998 Annual Report
Grand Challenge Projects
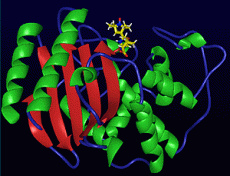
A model of the Michaelis complex for the TEM-1/penicillin system from molecular dynamics simulations.
Investigators: P. A. Bash, Northwestern University Medical School and M. Karplus, Harvard University
Research Objectives
A guiding principle of molecular biology is that the structure of a biomolecule defines its function. This principle is especially true in the case of the protein molecules known as enzymes, which serve as highly specific and extraordinarily efficient catalysts of biochemical reactions. Despite the growing availability of the atomic structures of enzymes, details of the chemical mechanisms employed by enzymes to achieve their catalytic prowess remain elusive.
The goal of this project is to develop a greater understanding of the mechanisms involved in enzyme catalysis. We are studying two different enzymes, one a protein, the other a nucleic acid. The protein enzyme that we are investigating is beta-lactamase, which is responsible for the efficient hydrolysis of antibiotic agents such as penicillin and cephalosporin. The nucleic acid enzyme we are investigating is the hammerhead ribozyme, which is a self-cleaving ribozyme suggested as a model for the primordial enzyme before proteins came into existence.
Computational Approach
The methods we use are based on the physical and chemical principles of statistical mechanics and quantum mechanics, and they are implemented in computational form using techniques from computational chemistry. To carry out our calculations, we used both the T3E and the J90 machines at NERSC. The molecular dynamics and quantum mechanical calculations were done on the T3E and J90 machines, respectively.
Accomplishments
We have developed a model of the Michaelis complex for the TEM-1/penicillin system from molecular dynamics simulations (see figure). We have also developed a model quantum and molecular mechanics (QM/MM) Hamiltonian for the beta-lactamase system, calibrating the Hamiltonian parameters against high levelab initiocalculations.
The catalytic mechanism of the hammerhead ribozyme is being investigated using a number of techniques that have been developed in our laboratories. An important goal is to determine the conformational flexibility of the active site of the ribozyme and the role that this may play in the catalytic mechanism. The new nucleic acid molecular mechanics force field in the CHARMM program will help us perform a classical molecular dynamics study of the hammerhead ribozyme.
Significance
The dynamic properties of proteins and nucleic acids are difficult to investigate experimentally, but they are essential for an understanding of their function. Computer simulations can provide the necessary insights, at an atomic level of detail, for a complete understanding of the relationship between biomolecular dynamics/structure and function.
For example, while the class of enzymes known as beta-lactamases are largely responsible for the increasing resistance of bacteria to antibiotics, the precise chemical resistance mechanism used by this enzyme is still unknown. Simulations are critical for further study of this mechanism.
Similarly, RNA was recently discovered to catalyze its own cleavage or that of other RNA molecules. The dual capacity of RNA to act as an information transfer molecule and an enzyme has led to speculation on the role of RNA in primordial life. Simulations involving the hammerhead ribozyme, the smallest of the catalytic RNAs, will be very useful in further investigations of RNA.
Publications
M. A. Cunningham and P. A. Bash, "Systematic procedure for the development of accurate QM/MM model Hamiltonians," inComputer Simulations in Biomolecular Systems, edited by W. F. Van Gunsteren, P. K. Weiner, and A. J. Wilkinson (Kluwer, Dordrecht, Neth., 1997), pp. 177-95.
M. A. Cunningham, R. E. Gillilan, and P. A. Bash, "Computational enzymology," Can. Chem. News 49, 9 (1997).
M. A. Cunningham and P. A. Bash, "Computational enzymology," Biochimie.79, 687-9 (1997).
About NERSC and Berkeley Lab
The National Energy Research Scientific Computing Center (NERSC) is a U.S. Department of Energy Office of Science User Facility that serves as the primary high performance computing center for scientific research sponsored by the Office of Science. Located at Lawrence Berkeley National Laboratory, NERSC serves almost 10,000 scientists at national laboratories and universities researching a wide range of problems in climate, fusion energy, materials science, physics, chemistry, computational biology, and other disciplines. Berkeley Lab is a DOE national laboratory located in Berkeley, California. It conducts unclassified scientific research and is managed by the University of California for the U.S. Department of Energy. »Learn more about computing sciences at Berkeley Lab.







